Although I’ve heard it’s really powerful, I’ve only used the Mac OS Alfred App for many years as a glorified app launcher or to trigger system commands like Sleep. I thought I’d look into what else I could get it to do.
The PediTools webtool I developed for the AAP 2022 hyperbilirubinemia guidelines can be accessed via an API.
How about analyzing bilirubin levels via a keyboard shortcut?
Google search came up with an thread from 2015 describing a bash script solution by Andrew Pepperrell. ChatGPT helped me translate the script from bash to Mac OS’s current zsh and handle optional arguments.
The solution worked nicely! For example, to load the page for:
- 40 week gestation newborn
- 24 hours age
- bilirubin 12 mg/dL
- and no risk factors (default)
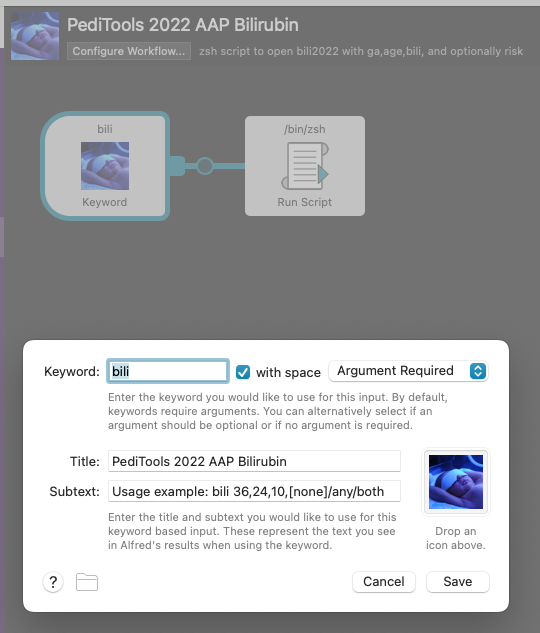
…the Alfred workflow is triggered by typing:
<Cmd-Space>bili 40,24,12<Return>(A fourth argument can be included of either any or both to reflect neurotoxicity risk factors – it defaults to none.)
That results in the zsh script generating and launching the following URL:
https://peditools.org/bili2022/api/?ga=40&age=24&bili=12&risk=none
Nice!
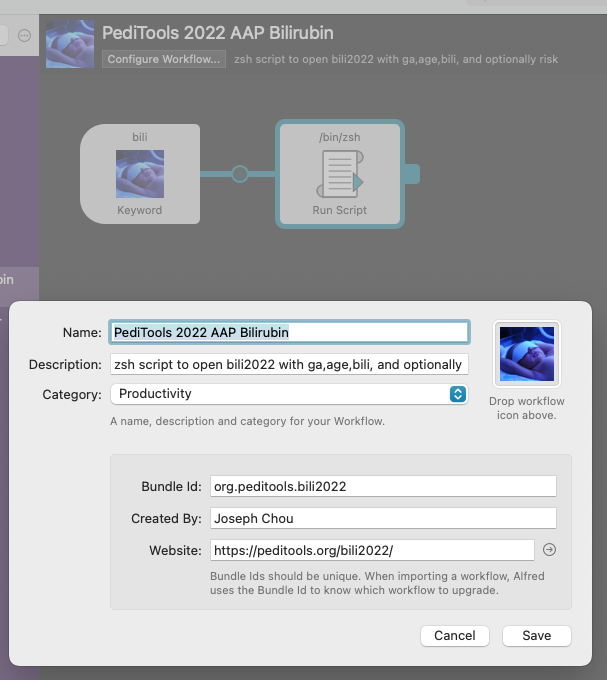
Screenshots of the Alfred Workflow setup:
Activation trigger
zsh script
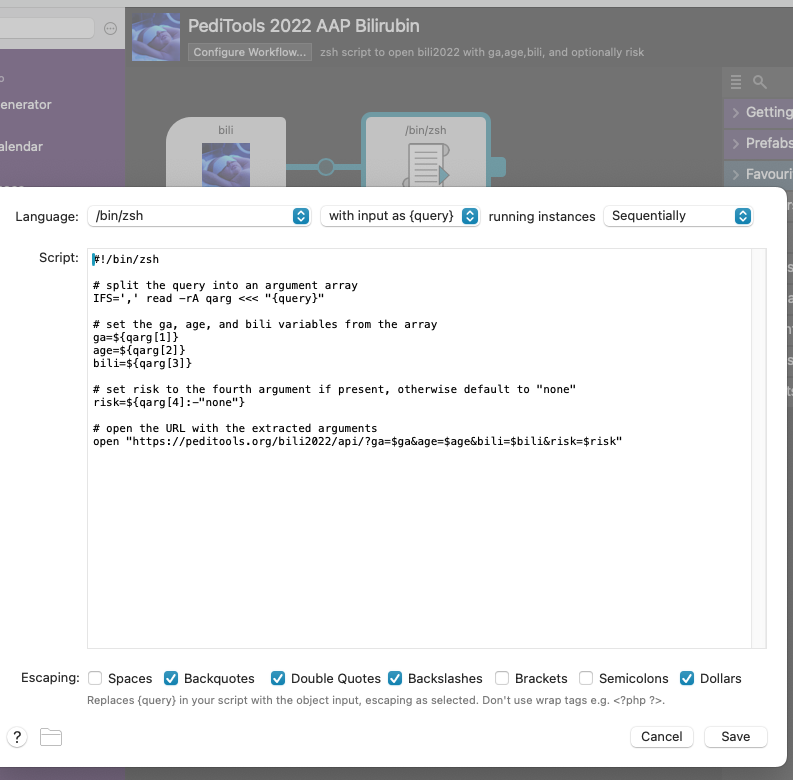
Heres the zsh script, for copy/paste:
#!/bin/zsh
# split the query into an argument array
IFS=',' read -rA qarg <<< "{query}"
# set the ga, age, and bili variables from the array
ga=${qarg[1]}
age=${qarg[2]}
bili=${qarg[3]}
# set risk to the fourth argument if present, otherwise default to "none"
risk=${qarg[4]:-"none"}
# open the URL with the extracted arguments
open "https://peditools.org/bili2022/api/?ga=$ga&age=$age&bili=$bili&risk=$risk"It was easier than I thought it would be. Makes me want to learn more about both Alfred and shell scripting.